Current Trends, Challenges, and Optimization Strategies in Bioinformatic Pipelines for Whole Genome Sequencing of Non-Model Species
Bioinformatic pipelines for whole genome resequencing in non-model species
Keywords:
Whole genome sequencing, Non-model species, Pipeline optimization, Variant calling tools, Reference genome bias, Population genomicsAbstract
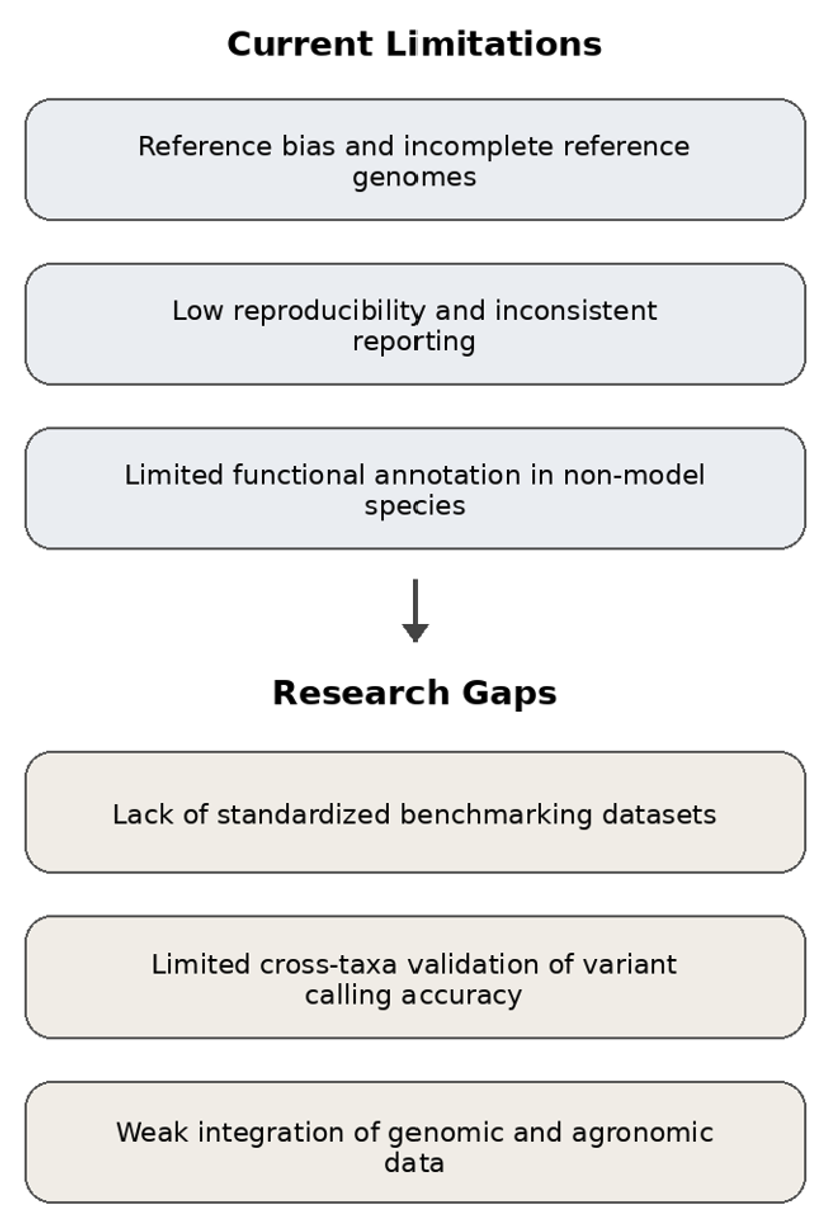
Whole genome sequencing (WGS) has become a central tool in evolutionary biology, conservation genetics, and agricultural genomics, enabling high-resolution analyses of genetic variation across diverse taxa. However, the application of WGS to non-model species presents substantial bioinformatic challenges, including incomplete or biased reference genomes, high levels of genetic diversity, variable sequencing depth, and limited computational resources. These constraints complicate pipeline design, variant discovery, and biological interpretation, particularly in agriculturally relevant systems where genomic outputs must be translated into practical outcomes.
This review critically examines current bioinformatic pipelines used for whole genome sequencing analyses in non-model species, with a focus on methodological trade-offs, sources of bias, and context-dependent optimization strategies. We synthesize recent advances in read processing, alignment and assembly approaches, variant calling frameworks, and functional annotation tools, and compare commonly used pipelines with respect to their suitability for non-model and agricultural applications. In addition, we highlight persistent limitations in benchmarking, reproducibility, and data integration, and discuss emerging trends such as long-read sequencing, pangenome frameworks, and machine learning-assisted pipeline optimization.
By integrating conceptual frameworks, comparative evaluations, and applied examples from crop, livestock, and pathogen genomics, this review provides practical guidance for designing robust and reproducible WGS bioinformatic workflows. The insights presented here aim to support informed methodological decision-making and to facilitate the effective translation of genomic data into agricultural improvement, conservation management, and biological discovery in non-model systems.
Downloads
Published
Issue
Section
License
Copyright (c) 2026 SciNex Journal of Advanced Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.




 SCINEX PUBLISHERS
SCINEX PUBLISHERS