Tryptic Digested Protein Transglutaminase Elicitor Can Improve Plant Physiological Parameters: A Computational Approach to Insight Molecular Interactions
Keywords:
Transglutaminase activity, Structural analysis, Pathogenicity protein, Functional annotation, TgaseAbstract
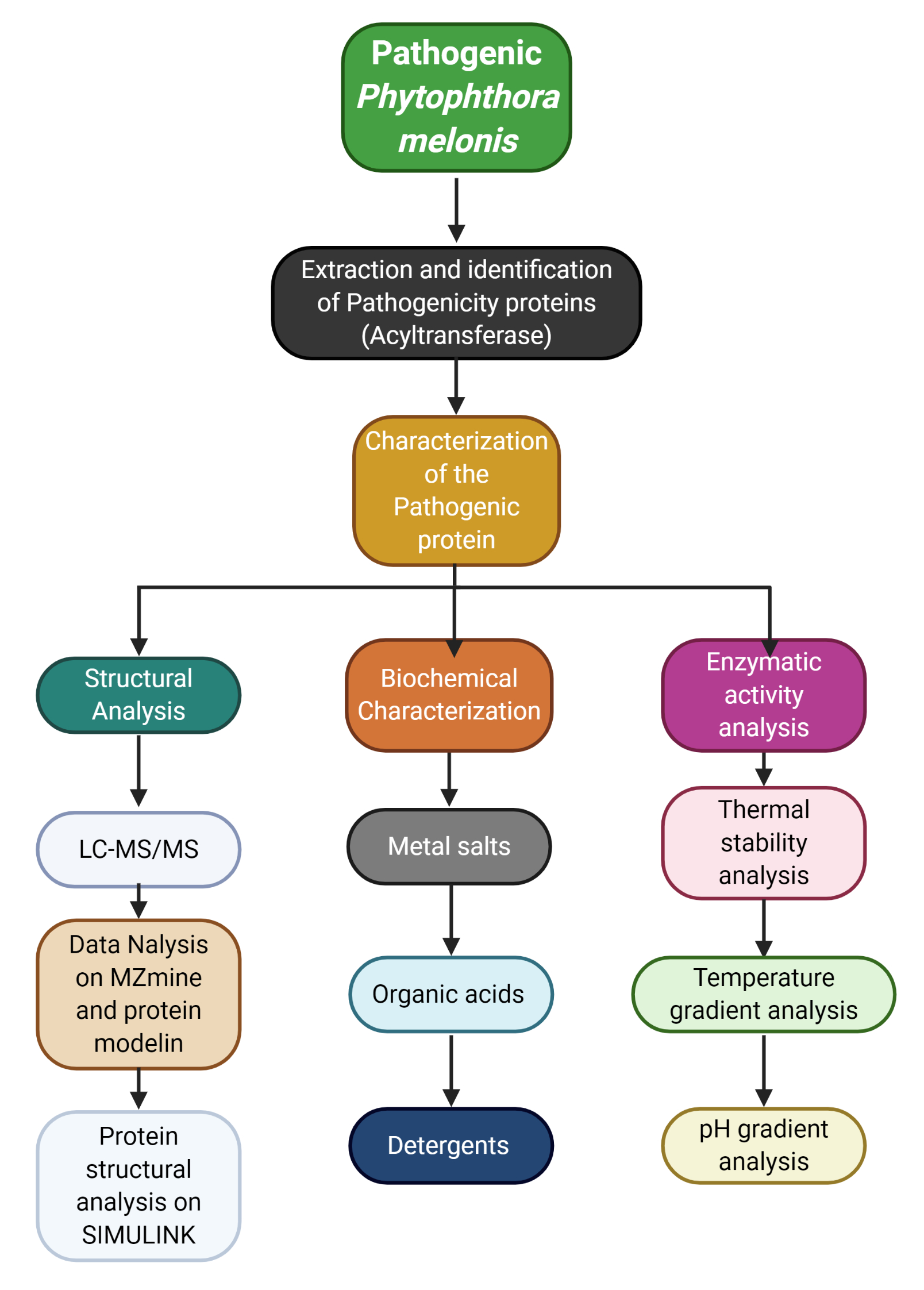
Phytophthora melonis causes horticultural crops' dieback, foot rot, root rot, crown rot, and blight. It possesses multiple pathogenicity factors, and transglutaminase is among the most effective pathogenicity tools of Phytophthora. We have isolated the pathogenic enzyme and applied its tryptically digested fragments to tomato plants to exploit effector proteins for the augmentation of plant defenses, physiological parameters, antioxidative machinery, and evapotranspiration. The results revealed that the treatment significantly increased agronomic traits, e.g., root length, shoot length, root biomass, shoot biomass, root dry mass, shoot dry mass, and leaf area, around 30% in each case. Furthermore, an increase of more than 10% was observed in the case of photosynthetic parameters. At the same time, the antioxidative machinery (enzymatic and biochemical) was more than 53% more efficient than the control plants. Similarly, the evapotranspiration and electrolyte leakage improved by more than 18%. Furthermore, the pathogenic enzyme underwent structural and functional characterization. The biochemical tolerance tests revealed the temperature of 37 °C and apH of 6.4 as the optimum circumstances of the enzyme. The Tween-80 application increased activity by 136.72%, although higher temperatures reduced the stability of the enzyme. The structural protein annotations exhibited 2 monomers, including A (260-620) and B(141-219). Elicitation of transglutaminase (TGase) and antifreeze attributes were revealed by monomer A and monomer B, respectively. Additionally, an association of 6 ligand molecules of 3-cyclohexyl-1-propylsulfonic acidic were observed to maintain the enzymatic activity. Overall, the protein was categorized as the freeze-resistant protein that was revealed in three genera, i.e., Plasmopara, Phytophthora, and Pythium, according to the biome distribution analysis. Whereas the protein was also present in some other taxonomic classes (i.e., archaea, and bacteria). The current study is a new approach to exploit the plant metabolic machinery for improved cellular defenses and metabolism. It will also help to interpret the supramolecular interactions between the pathogen and its host, especially in a pathogen from oomycetes.
Downloads
Published
Issue
Section
License
Copyright (c) 2026 SciNex Journal of Advanced Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.




 SCINEX PUBLISHERS
SCINEX PUBLISHERS